Proteomics and Its Clinical Application
How proteomics offers opportunities for understanding complex diseases, like cancer.
How proteomics offers opportunities for understanding complex diseases, like cancer.
Proteomics is the large-scale study of proteins, particularly their structures and functions. In its clinical application, proteomics encompasses the work of applying protein-based discoveries and technologies to improve patient care.
Following the sequencing and mapping of the human genome roughly two decades ago, an era of increased interest and investment into proteomic research has led to dramatic advances in the techniques and technologies powering proteomics.
These achievements have since enabled the rapid, accurate identification of key molecules and assays within the proteome, presenting new opportunities for augmenting and enhancing genomic tools to further enhance clinical diagnostic and treatment options.
While the work required to understand proteins on a grand scale and bring about radical change to clinical care continues to surprise in its complexity and required diligence, tangible accomplishments in the development of clinical proteomic tests are already being utilized by physicians to overcome the immense and growing challenge of diagnosing diseases and charting a more informed path of care for patients across the country.
This guide offers an overview of proteomics within the frame of its clinical application. We’ll explore why proteomics is important for understanding and treating diseases, how it complements and expands upon achievements in applied genomics, and which tools and techniques present both current and future solutions for clinicians.
Similar to the remarkable abilities enabled through achievements in genomics (the sequencing of the human genome, in particular), the characterization of proteins achieved through proteomics is already providing deep, clinically-useful insight into how diseases function and how best to treat them at an individual level.
Understanding how proteomics provides this clinical value requires a basic understanding of the nature of human disease. While our genomes serve as the blueprints for protein production, those proteins go on to perform essential functions in the body. In abnormal disease states, gene mutations can cause these proteins to become deregulated and defective––giving rise to major health conditions, including heart and muscle diseases and cancer.
Proteomes, the entire sets of proteins present in a cell or tissue, are more complex and dynamic than genomes. This is because each gene can potentially bring about more than one protein.
Making matters even more complex, the abundance of each protein is regulated, and proteins typically become chemically processed and modified––often engaging in complex interactions with other proteins or small molecules (e.g. metabolites, lipids, metal ions and co-factors).
Technological advancements have made genomic DNA and RNA sequence information readily accessible. However, the structures, functions, drug interactions, and regulation of proteins are not predictable based on genomic sequences.
This is where proteomic research and development offers a massive opportunity for understanding these deregulated, disease-causing proteins, thereby creating new opportunities for developing novel tools clinicians can utilize for more effective diagnosis and treatment.
In general, parsing the question of “why proteomics?”––specifically in clinical research and development––can be distilled into a couple major points that are closely related to one another:
Interpreting the cancer ecosystem offers a clear example of how genomic and proteomic data combine to provide clinically valuable intelligence. While genomic data uncovers the potential genomic drivers of cancer (tumors in particular), proteomic data profiles the patient’s (or host’s) immune response to the cancer. Both are critical elements for interpreting the real-time complexity of the cancer disease state or cancer ecosystem.
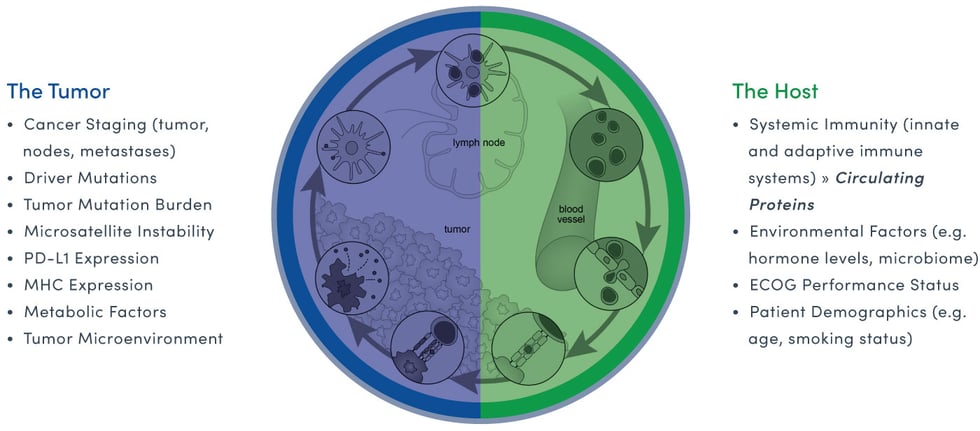
The easiest way to understand why it’s important to study the genome and the proteome for the best clinical outcome is through the simple analogy of a caterpillar morphing into a butterfly. Both have identical genomes but vastly different proteomes. This enables the caterpillar to crawl from place to place and the butterfly, the ability to fly.
Similarly, cancer-cell to host-cell interactions create a new, complex environment, that when properly understood, can directly inform clinical decisions based on the real-time status of the cancer ecosystem.
The demonstrated weaknesses of applied genomics have been well established, but so too have the strengths of proteomics as a means of overcoming them.
The global gene expression information revealed through genomics is absolutely critical for developing the molecular markers clinicians use to distinguish different disease states and make useful correlations from clinical data.
... once a deregulated gene or pathway is identified, it must be followed up by more focused research to further investigate the biological and clinical implications of the deregulation. While the results in the molecular ignatures are robust, the changes in individual gene expression do not always translate into the changes in its protein or protein modification. Proteins from these deregulated genes being the functional excecutioners in the cells, proteomics is the most logical next step in our understanding of biological processes. (Chung at al, 2007)
But genomic molecular markers can only go so far in painting a complete picture of the biological processes making up a patient’s individual disease state (and thus providing a way to better understand and treat it).
In short, the relevant limitation of genomics, in its current state, is its inability to capture the important contextual factors that impact gene expression. As revealed by Chung et al, changes in how an individual's genes are expressed do not always translate into alterations in its proteins or their modifications.
Since it’s the proteins from these deregulated genes that act as the “functional executioners” within the cells, proteomics provides a viable pathway for developing new clinical tools that complement and augment genomic tools precisely at their weakest points. Put more simply, today’s proteomic research and subsequent transition to clinical tool development offer a set of keys for unlocking many of the doors genomics alone cannot open. Behind them lie opportunities for developing clinical tools that, until now, have remained out of reach.
To understand the state of a complex disease like cancer at a certain point in time, it’s important to examine more than just the tumor tissue. Proteins in a patient’s blood form a synergistic relationship with both cancer tissue proteins and various factors of the individual's biology, including their immune system. This interaction affects the state of their blood-based protein content, which can then be interpreted through advanced proteomic tools to reveal the dynamic change of the disease state.
As we’ll explore in more detail in a moment, by characterizing this affected protein content through innovative proteomic tools and techniques, valuable cancer biomarkers can be identified and used to provide clinically-valuable insight into the real-time status of a patient’s health.
These tools and technologies have garnered significant interest for pharmaceutical and diagnostic development due to the easily accessible nature of blood as a source of rich protein content that lends itself to convenient clinical application.
As summarized by E. Maes et al, proteomic clinical tools focused on advanced blood protein analysis provide a number of generalized, clinically-relevant advantages that complement and expand on the capabilities of genomic technologies:
While many of these advantages have been realized for years, the techniques that power proteomic analysis have only more recently crossed the threshold into clinical viability thanks to technological improvements that make such tools accessible in a clinical setting.
Mass spectrometry-based proteomic technologies, for example, have become faster and more sensitive––analyzing the thousands of proteins from a clinical sample and generating tremendous amounts of information used to characterize the underlying biology within a given disease state.
Here, the “hand-off” from genomics to proteomics (and the difference between tissue and blood protein analysis) becomes clear and meaningful. Armed with proteomic tools and informed by genomic insights, clinicians can now untangle and understand how biological systems are regulating the processes that directly impact the nature of growth and aggressiveness in the tumor.
The real-world application of these sophisticated proteomic techniques, as conveniently collected and summarized by Carl A. K. Borrebaeck in a 2017 Nature Reviews Cancer article, is already having a meaningful clinical impact in evaluating specific disease states, which has only continued to progress and accelerate.
The difference between tissue and blood proteins, and what each can tell us, is especially salient in the search for predictive and prognostic cancer biomarkers.
In the past, precision oncologists focused their search for these biomarkers on those arising directly from malignant tumor tissue cells. These can serve to differentiate cancer cells from healthy cells, distinguish more aggressive cancer cell types from less aggressive ones, and identify weaknesses such as “gene addictions” that could be exploited to disrupt cancer progression.
While these insights shed valuable light on how complex disease states function, their limitations, specifically in clinical applications, have prompted a shift in the hunt for biomarkers.
Increasingly, biological signals have been identified that arise not only from the tumor cells themselves but from the patient’s (or host’s) own physiology.
Research into the biological microenvironment surrounding the tumor has revealed a role for “bystander” cells, such as immune cells and stromal cells, which interact with tumor cells. These have given rise to biomarkers that, when paired with sophisticated analytical tools, sharpen the insights available to clinicians in delivering care, specifically within the context of today’s most promising routes of therapy.
Immunotherapy, in particular, has reinforced the role of the components of the immune system as either proponents or antagonists of the cancer-immunity cycle and therefore has identified them as useful biomarkers of patient outcome in the context of immunotherapy. (Chen and Mellman, 2013)
Even the microbiome has been identified as a contributor to the cancer-immunity cycle and potential source of biomarkers. (Vargas, 2016) Together with our modern understanding of cancer as a systemic disease, this has expanded the biomarker search space from the tumor to its microenvironment and beyond within the host physiology.
These biomarkers may be reflective of the state of the tumor microenvironment or the larger host biology and its interaction with the tumor. And while smaller biomarker panel sizes and tumor-centric markers are often more mechanistic and targetable, host factors are often easier to sample and, when paired with epidemiological outcome data, yield predictive and prognostic insights with great potential for clinical utility.
Since its emergence and subsequent acceleration following the completion of the Human Genome Project, proteomics has given rise to a new set of methodologies, techniques, and technologies to aid in understanding, detecting, and treating diseases like cancer by characterizing the protein networks that both control and are controlled by the information encoded by the genome. (Srinivas et al, 2002)
All of these tools and methods, as Srinivas et al explain, must contend with challenges unique to the study of proteins. These include the inherent complexity of protein regulation, difficulties in capturing protein targets given the specific determinants of their behavior, and the task of detecting low-abundance proteins given that the dynamic ranges of proteins in biological systems can reach parts per million or lower.
Similar to the international bodies that have organized to overcome complex challenges in genomics, proteomic networks like The Human Cancer Proteome Project and Human Proteome Organization are fostering cooperation and collaboration to develop and deploy better proteomic-powered tools around the world.
These networks have created multidisciplinary groups of cancer proteome scientists, pathologists, and other clinicians to standardize practices and protocols for critical research tasks, such as tissue collection, reporting, and data sharing. This collaboration is enabling the large-scale meta- and pan-cancer analyses needed to interrogate cancer at the proteome level and integrate knowledge to improve clinical decision-making and spark new clinical and translational research.
On the ground level, these efforts have produced a number of technological platforms and methodologies currently utilized for proteomic discovery and subsequent analysis. We’ve focused our attention on two that are particularly relevant to clinical application and, as Belczacka et al show through collected studies, are among the most widely applied discovery methods in use today: Liquid Chromatography Mass Spectrometry (LC-MS) and Matrix-Assisted Laser Desorption/Ionization Time of Flight (MALDI-ToF).
Chung et al succinctly compare these two strategies for identifying and profiling proteins by mass spectrometry as “bottom-up” and “top-down” proteomics.
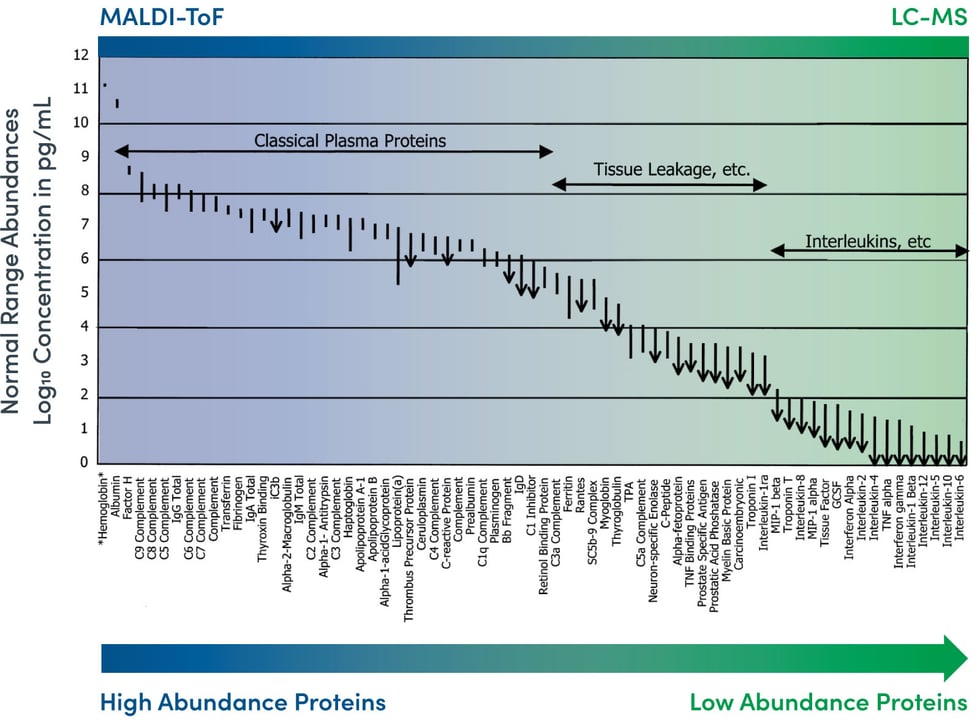
Image Source: N.L. Anderson and N.G. Anderson. The Human Plasma Proteome. Molecular and Cellular Proteomics (2002); 1:845-867.
In the “bottom-up” approach using LC-MS, a sample containing a simple or complex mixture of proteins is digested with a protease. Subsequent separation of all of the resulting peptides is performed using liquid chromatography.
In the “top-down” approach using MALDI-ToF, soft ionization allows for the optimal analyzation of larger organic molecules like peptides, lipids, and saccharides without fragmenting or decomposing them––a common drawback of traditional techniques.
Multiple Reaction Monitoring Mass Spectrometry (MRM-MS) is a technique that quantifies predefined highly-multiplexed sets of proteins with high sensitivity, specificity, and reproducibility.
As explored in-depth by Yates et al, the popularity of LC-MS/MS stems from its effectiveness in large-scale protein profiling of highly complex samples, providing the high-resolution proteomics data that lends itself to clinical use.
Belczacka et al note that another major advantage of LC-MS/MS is its ease of use and the fact that its protocols for sample preparation and analysis have been extensively tested and are available, making for a “straightforward” application on a technical level.
This method is well suited for the detection of abundance changes in a large set of proteins in a single analysis. Changes in protein abundance can be used to stratify disease states, optimize therapeutic selection, and determine prognosis.
LC-MS when applied in this clinical application, offers a number of key benefits as well:
MRM-MS is ideal for researchers asking, “What happens to my proteins of interest?” Since there is no need for expensive and time-consuming antibody development, this approach offers significant time and cost savings over antibody-based methods. MRM-MS has significant advantages over immunoassays, in particular in multiplexing capability, specificity, and eliminating the need for expensive and time-consuming generation of custom reagents. (Kearney et al, 2018)
Matrix Assisted Laser Desorption/Ionization Time of Flight (MALDI-ToF) mass spectrometry is one technique used to interrogate the proteome––including post-translational modifications and splice variants––without the need to identify specific proteins in advance.
MALDI-ToF mass spectrometry, when applied in this clinical application, offers a number of key benefits that enhance its convenience and effectiveness:
Here at Biodesix, we develop multivariate proteomic tests that can measure protein abundance to gain a real-time comprehensive view of a patient’s changing disease state and immune profile. Blood-based proteomic analyses reflect innate or adaptive immune responses that may affect clinical outcomes on immunotherapy and other treatments
Perhaps the best way to understand what’s motivating the research and development in applied proteomics is to understand the clinical questions it serves to answer. One of these critical questions speaks to the broad goals of treating complex disease states, and is one we’ve touched on throughout this piece:
Can the information contained in blood-based protein biomarker signatures (as opposed to individual biomarkers) provide the long-sought accuracy in cancer diagnostics needed for more precise, evidence-based options for managing complex disease states like cancer?
The ability to make informed predictions about clinical therapies and disease progression at the individual level is arguably among the most important practical advantages for care providers today. As we explore below, new diagnostics are providing clinicians the insights they need to deliver higher quality care at a time when looming public health concerns are colliding with an increasingly complicated healthcare environment to boldly underscore the need for greater efficiency and efficacy.
Recently expanded Medicare coverage for lung cancer screening is estimated to roughly double the number of lung nodules that will require evaluation and management in the United States each year, from the roughly 1.57 million currently detected incidentally to roughly three million.
This dramatic expansion is poised to create an enormous challenge for physicians tasked with estimating the probability of cancer and choosing the appropriate management path for all of these patients.
Nodify XL2™ is a blood-based laboratory developed test (LDT) that enables physicians to access a classifier that integrates circulating proteins and clinical parameters that may add benefit in the management of lung nodules by providing additional assurance to patients with likely benign nodules to follow an imaging surveillance path, while decreasing the risks and costs of otherwise avoidable diagnostic and invasive procedures.
Nodify XL2 works as a “Rule-Out” test, identifying patients with benign lung nodules who can avoid unnecessary invasive procedures up to and including surgical resection. The test measures the relative abundance of two peptides contained within circulating plasma proteins (LG3BP and C163A) as assayed using mass spectrometry. The native proteins from which the peptides are derived, have been associated with an inflammatory response to cancer.
These protein measurements are then integrated with five clinical risk factors (age, smoking status, nodule diameter, nodule spiculation status and nodule location) to arrive at a numerical risk score between zero and one.
Among all patients diagnosed with lung cancer, 70-80% are diagnosed specifically with advanced non-small cell lung cancer (NSCLC). In 84% of cases, NSCLC is diagnosed at an advanced stage when it has largely become incurable. As a result, improved functional status, improved quality of life, and improved overall survival are significant factors to consider for treatment decisions.
Achieving these treatment goals requires fully characterizing the patients’ disease state, which is defined by both tumor characteristics (such as size, location, anatomy, mutational profile) and the patient’s immunological response to the tumor (host response or host biology).
The VeriStrat® test is the only clinically validated test available for profiling this critical host response. VeriStrat is a blood-based proteomic test that does not require tissue or additional surgical biopsy. Instead, it utilizes liquid biopsy and proteomic profiling through a mass spectrometry (MALDI-ToF) methodology to identify a chronic inflammatory immune response indicative of aggressive disease in patients with advanced NSCLC, allowing for fast turnaround time to results in 72 hours or less.
With a binary result of VeriStrat Good (VS Good) or VeriStrat Poor (VS Poor), the test identifies patients with an aggressive immune component to their disease state (VS Poor patients) who are unlikely to respond to standard of care therapies and may benefit from investigational therapies in clinical trials. On the other hand, patients with a VS Good disease state are significantly better candidates for standard of care therapies, including immunotherapy and more aggressive combination therapies. VS Good patients have an overall survival that is twice as long as those patients who test VS Poor. The VeriStrat test creates an opportunity to improve patient QOL and increase survival outcomes without requiring the use of tissue ––significantly improving cancer care.
The mission to understand and manage complex diseases like cancer presents some of the most significant challenges in clinical research and development. Recent innovations in precision diagnostics, enabled through proteomics, offers practical, evidence-based opportunities for detecting and treating these diseases by deciphering the rich, clinically-valuable information contained within the proteome, that until recently, has remained frustratingly out of reach.
Here at Biodesix, we’re working to develop and deploy advanced diagnostic solutions to improve the standard of personalized care for patients by equipping clinicians with the right information at the right time. Learn more about our blood-based tests and how they leverage genomics and proteomics to uncover individualized insights about tumor biology and patient immune responses.
A leading data-driven diagnostic solutions company helping answer critical clinical questions faced by physicians, researchers, and biopharmaceutical companies, with a primary focus in lung disease.
CORPORATE HEADQUARTERS
919 W. Dillon Rd. | Louisville,
CO 80027
Phone (303) 417-0500
Fax (866) 432-3338
© Copyright 2024 Biodesix.
All Rights Reserved.
Genomics and proteomics: Emerging technologies in clinical cancer research
Proteomics in cancer research: Are we ready for clinical practice?
Precision diagnostics: moving toward protein biomarker signatures of clinical utility in cancer.
Prostate cancer screening in men aged 50-69 years (STHLM3): a prospective population-based diagnostic study
Biomarker development in the precision medicine era: lung cancer as a case study
Large-scale protein identification using mass spectrometry
The building blocks of successful translation of proteomics to the clinic
Proteomics biomarkers for solid tumors: current status and future prospects